Restriction Fragment Length Polymorphism (RFLP)

Genetics is an extensive research field due to genetic diversity among populations, between individuals, and even between species and the discoveries of genetic diseases and its cure. This makes the analysis of genetic diversity extremely important. As we already know, this is possible due to recent discoveries of certain tools used in genetics. Restriction Fragment Length Polymorphism is one of the tools and is being used as a genetic marker. These are highly reproducible hybridization-based markers. Improved restriction fragment length polymorphism uses PCR and is also known as CAPS.
A must read- Microinjection: Method of gene transfer (mybiologydictionary.com)
Table of Contents
RESTRICTION FRAGMENT LENGTH POLYMORPHISM AS GENETIC MARKERS
Genetic markers or molecular markers are DNA fragment associated with a genome that does not take part in coding or translation. Being the noncoding segment, these markers are stable.
Thus, due to their stability, cost-effectiveness, and ease of use, these markers provide an immensely popular tool for a variety of applications including genome mapping, gene tagging, phylogenetic analysis, and forensic investigations. The most common molecular markers applied in genetics are RFLPs, SSR, STR, ISSR, RAPDs, AFLPs,` SSCPs, CAPS, etc.
FIRST GENERATION MARKERS
The molecular markers that are based on Southern Blotting are first-generation markers like RFLPs. Thus, most of the RFLP fragments can be detected and analyzed using nucleic acid probes that are radioactively labelled. The main advantage of RFLP analysis over PCR-based markers is that no prior sequence information nor oligonucleotide synthesis is required.
RESTRICTION FRAGMENT LENGTH POLYMORPHISM
Restriction Fragment Length Polymorphism as the genetic marker is a technique that uses the detection of genetic polymorphisms that results in variations of restriction fragment patterns of a different region in the genome.
RFLP is an abbreviation that stands for restriction fragment length polymorphism. The reason for assigning such an abbreviation can be understood if we focus little on the technique itself, as it uses molecular scissors (restriction enzymes) to cut DNA into pieces (fragments) and detect the changes (polymorphism) in size (length) of the specific repeat sequences.
SOURCE of RFLPs: Satellite DNA
Only about 1 per cent of eukaryotic DNA is made up of protein-coding genes and the other 99 per cent is non-coding. Noncoding does not provide instructions for making proteins. There are certain locations in a non-coding region where a short nucleotide sequence is repeated many times and is called repetitive DNA. This repetitive DNA forms small peaks during density gradient centrifugation, hence referred to as satellite DNA.
These short repetitive nucleotide sequences are organized as a tandem repeat called a variable number tandem repeat (VNTR). These can be found on many chromosomes and often show variations in length between individuals. VNTRs are an important source of restriction fragment length polymorphism as a genetic marker. So, VNTRs can be extracted with restriction enzymes and analyzed by an RFLP marker.
GENETIC POLYMORPHISM
Genetic polymorphism is a difference in genomic sequences among individuals that is caused due to mutations. Thus, mutations in DNA generate polymorphic DNA molecules that vary from person to person and are unique for every individual. To detect these changes, certain genetic markers like the RFLP marker are used.
RFLP takes advantage of the polymorphism in the genetic code of individuals and the pattern fragment sizes are tested.
RFLP as Co-dominant Marker
Molecular markers are either dominant or co-dominant.
Dominant markers are unable to detect allelic variations or identify only a single dominant allele. Whereas, Co-dominant markers are able to detect heterozygosity or both the alleles of a diploid organism.
Restriction fragment length polymorphism as the genetic marker is co-dominant and highly locus-specific w makes it usable in DNA fingerprinting, expressing both parental RFLP markers for the locus regardless of it being recessive or dominant when co-occurring in an individual. RFLP genotyping has high reproducibility.
HYBRIDIZATION PRINCIPLE
RFLP uses hybridization which states that a single-stranded DNA or RNA molecule of a defined sequence can base-pair to a second DNA or RNA molecule that contains a complementary sequence, with the stability of the hybrid depending on the extent of base pairing that occurs. Restriction Fragment Length Polymorphism as the genetic marker is a labelled DNA sequence that hybridizes with one or more fragments of the digested DNA sample after they are separated by gel electrophoresis.
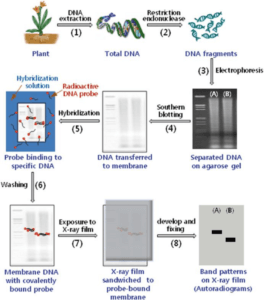
PROCESS OF RESTRICTION FRAGMENT LENGTH POLYMORPHISM
Advances in Veterinary Sciences- Sidharth Mishra, Chinmoy Mishra, Subhash Taraphder.
ISOLATION OF DNA
DNA can be extracted from saliva, semen, blood, hair bulb, or any other cell of the body. In order to cut DNA into fragments, it needs to be in pure form. This is achieved by treating the cell with enzymes to remove other macromolecules.
DNA FRAGMENTATION
Purified DNA is then subjected to restriction endonucleases. Restriction enzymes are the molecular scissors that recognize specific sequences in DNA that are restriction endonuclease enzyme recognition sites and cleave it. These fragments are of different lengths in different individuals due to polymorphism of DNA. Thus, any restriction fragment length polymorphism as the genetic marker is unique to specific restriction enzymes.
GEL ELECTROPHORESIS
The overall charge on DNA is negative, so, the restriction fragments we receive after endonuclease action is negatively charged. Now, we need to perform gel electrophoresis for the separation of fragments. For the same, the DNA ladder is loaded in the first well, and samples of fragmented DNA are poured into the other wells towards negative ends and made to run through a gel under an applied electric field. The DNA ladder is just like a scale or DNA of known sequences and molecular weight with which we compare the size of sample DNA after the run.
Being negatively charged, DNA fragments start to move towards the positive electrode, the smaller fragments move faster than the larger fragments, thus separating the DNA samples into distinct bands.
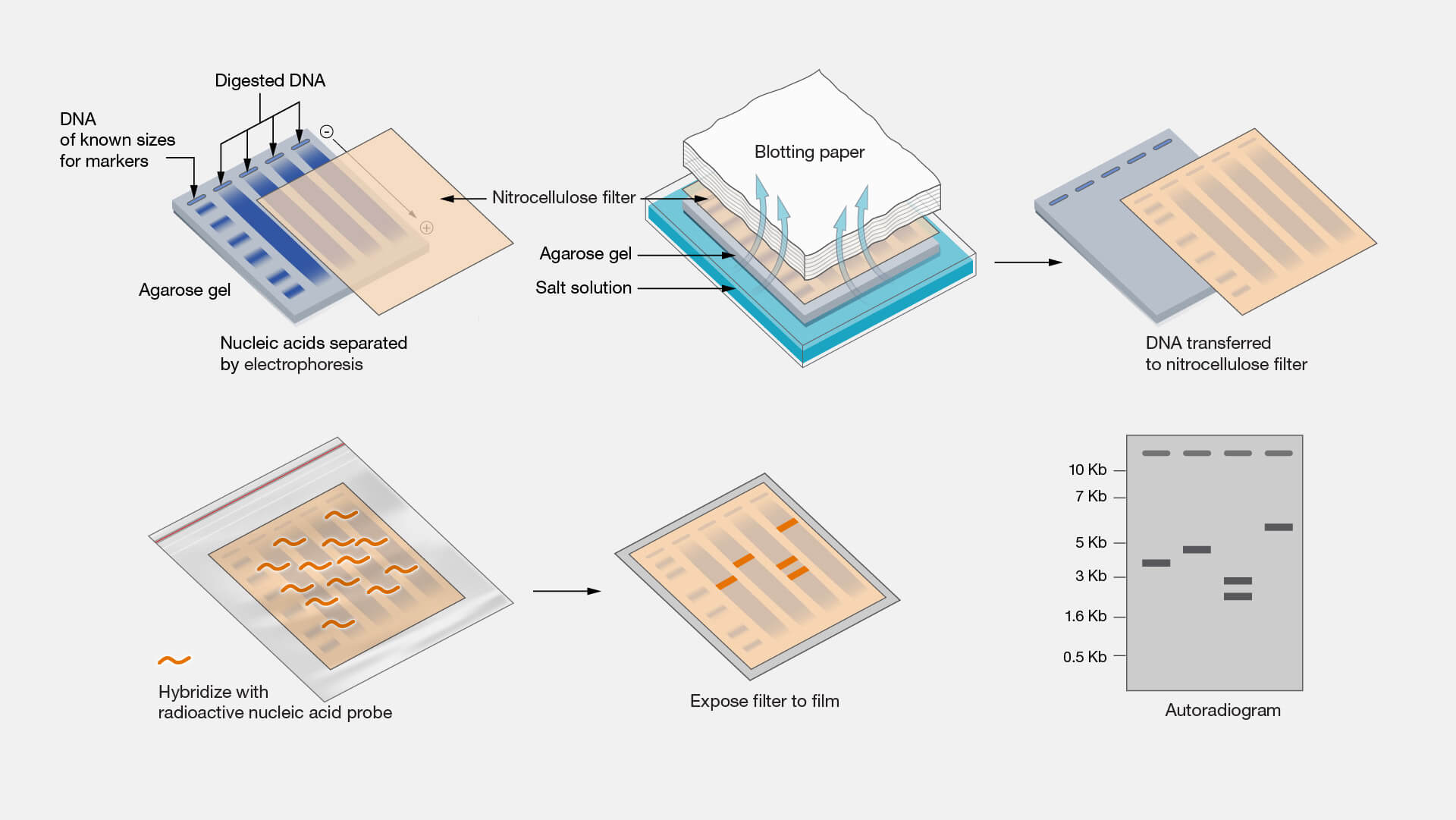
DETECTION THROUGH SOUTHERN BLOTTING
Here, the agarose gel with separated DNA fragments from electrophoresis is treated with strong alkali to denature it. This denaturing is necessary for the generation of single-stranded DNA so that it easily binds to its complementary probe.
Now, the southern blotting is performed that transfers, immobilize, and detects target DNA fragments onto the nitrocellulose membrane. The gel is placed on the filter paper wick and a nitrocellulose membrane is placed above which there are paper tissues. Transfer buffer is drawn through the gel by capillary action, and the nucleic acid fragments are transferred out of the gel onto the nitrocellulose membrane.
IDENTIFICATION OF SEQUENCE OF INTEREST
UV radiations are applied to the membrane to immobilize DNA fragments onto nitrocellulose by cross-linking. After the fixation step, the membrane is placed in a solution of labeled (radioactive or non-radioactive) RNA. This labeled nucleic acid is used to detect and locate the complementary sequence, called a probe.
https://www.genome.gov/genetics-glossary/Southern-Blot
The probe is allowed to hybridize with its complementary single-stranded target DNA sequences on the membrane. After the hybridization, the membrane is washed extensively to remove the non-specifically bound probe. If the probe is radioisotope labelled, then the membrane is exposed to photographic film. If the probe is non-isotopically labelled, the membrane may be treated with the chemiluminescent substrate to detect the labelled probe, and then exposed to photographic film.
Here, it is important to notice that any restriction fragment length polymorphism as a genetic marker is unique to the nucleic acid probe.
APPLICATIONS of RESTRICTION FRAGMENT LENGTH POLYMORPHISM AS GENETIC
Restriction Fragment Length Polymorphism Detection of Diseases: GENETIC MAPPING
RFLP marker is used to determine diseases or particular mutations in the genome and also differential diagnosis of hereditary diseases. This technique is very useful in determining the disease status of an individual that as Huntington’s chorea, cystic fibrosis, sickle cell anaemia, and so on.
FORENSIC INVESTIGATIONS
This technique is useful in solving criminal cases, paternity cases, and forensic sciences. DNA fingerprinting eventually discriminates the genome at its VNTR probes and helps in gene analysis.
AGRICULTURE
Any desired gene of interest can be isolated from the genome pool of a plant to produce certain disease-resistant, drought-resistant, and pests-resistant species.
To summarize, RFLP is a very useful technique in the process of Biotechnology. Keep reading to find out more!
Team MBD
Watch-RFLP | Restriction Fragment Length Polymorphism – YouTube


